Further Reading
Expanding the scope of protein biosynthesis
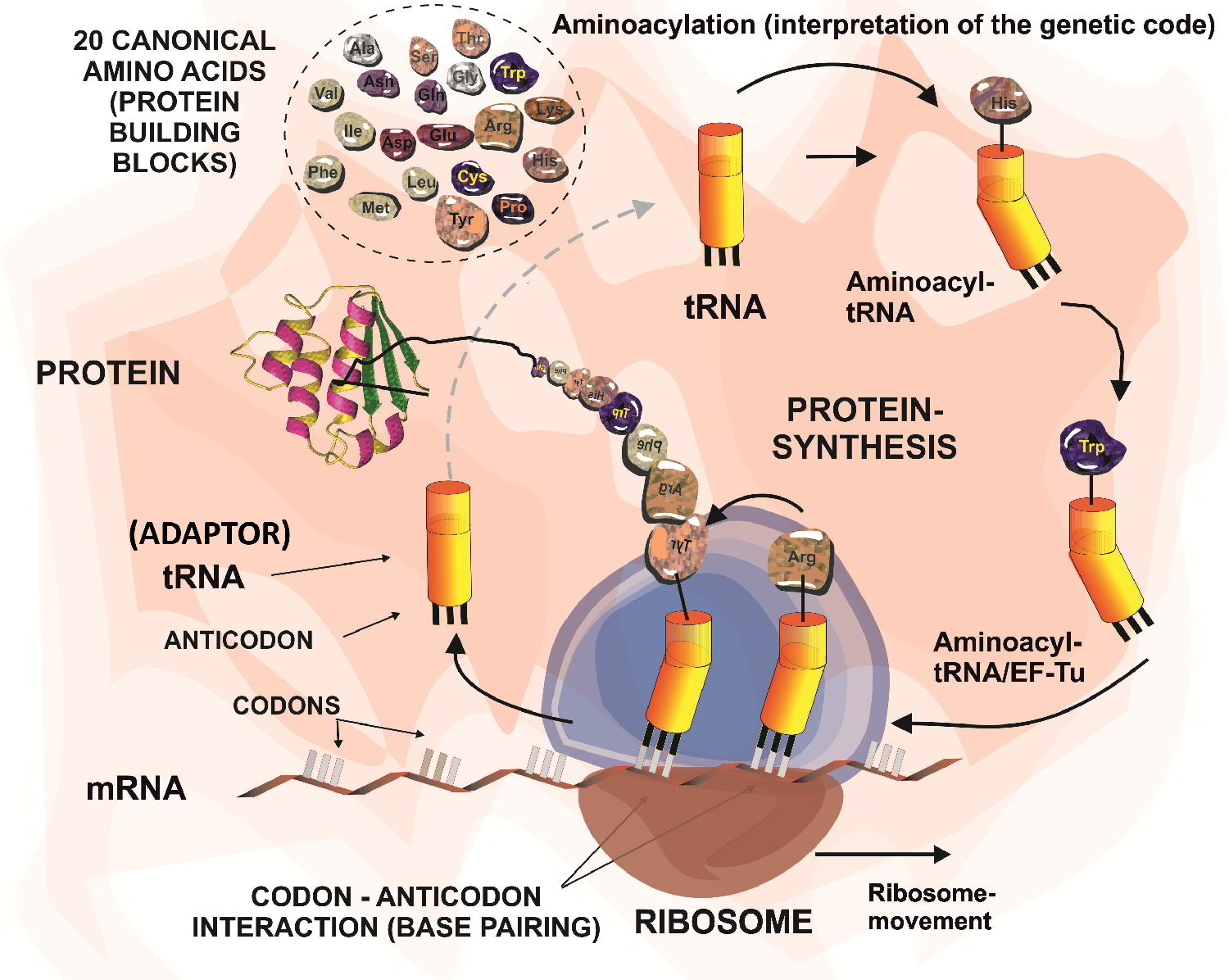
The aim of our research is to exchange, reduce or expand the number of amino acids as basic building blocks for protein synthesis, metabolism, cell structures and processes. This requires, first and foremost, the complex and radical bio-engineering with a focus on reprogramming of translation machinery by changing the coding rules, capacities and features of the genetic code.
Keep in mind following: no life forms are known to introduce amino acids into proteins beyond 20 (+2) canonical amino acids standard repertoire. However, the number of potentially novel building blocks for protein biosynthesis is virtually unlimited. This is plausible since in vitro works have demonstrated that creation of a totally new genetic code set is possible.
Therefore, we are very interested in studying the translational activity for the many non-canonical amino acids of natural or synthetic origin. Usually, code engineering is accomplished by use of appropriate metabolic prototypes (e.g. auxotrophies) or by direct engineering of the components of the ribosomal translation machinery (primarily aminoacyl-tRNA synthetases and tRNAs) or by a combination of both.
Genetic code engineering is aimed to explore the impact of new chemical functionalities in proteins, proteomes and whole cells. Namely, organic chemistry can provide a great diversity of non-canonical amino acids and unnatural cofactors that can be introduced into microorganisms as building blocks by experimental evolution.
Current efforts to “expand” the genetic code are in fact sophisticated in vivo protein labelling techniques primarily aimed at modifying the composition of individual proteins expressed in the frame of recombinant DNA technology. Thereby specific protein modification(s) is /are achieved by temporarily changing [re-assigning] decoding events involving only one target gene. This is the mainstream trend in this research area. However, a much more difficult task is to change (or redesign) the decoding events that are relevant to all cellular genes, resulting in a new amino acid insertion into the entire proteome.
This would require complex changes in the cell metabolic, regulatory and signaling networks in the target cells under the condition of experimentally designed long-term cultivation experiments. Clearly, this is most promising avenue towards the creation of bio-contained organism (e.g. bio-containment with genetic firewall against horizontal gene transfer). Such platforms should enable us to increase the chemical complexity, starting with a simple chemical analogues and surrogates of canonical amino acids or nucleobases, to chemically more distant and even radically different alternatives.
Despite rapid recent progress, it is not yet possible to completely alienate an organism that would use and maintain different genetic code associations permanently
Reading for more in-depth coverage of the subject
- Koch, N.G., Budisa, N. (2024) Evolution of pyrrolysyl-tRNA synthetase: from methanogenesis to genetic code expansion. Chem. Rev. 124(16): 9580–9608; doi: 10.1021/acs.chemrev.4c00031
- Hoesl, M. G., Budisa, N. (2011) Expanding and Engineering the Genetic Code in a Single Expression Experiment. ChemBioChem 12, 552-555; doi: 10.1002/cbic.201000586
- Wiltschi, B. and Budisa, N. (2007) Natural history and experimental evolution of the genetic code. Appl. Microbiol. Biotechnol. 74, 739 -753; doi: 10.1007/s00253-006-0823-6
Our Troika: what are we particularly interested in?
Our research focuses on studies and engineering of the genetic code using three canonical amino acids ("troika"):
- Proline (Pro, P)
- Methionine (Met, M)
- Tryptophan (Trp, W).
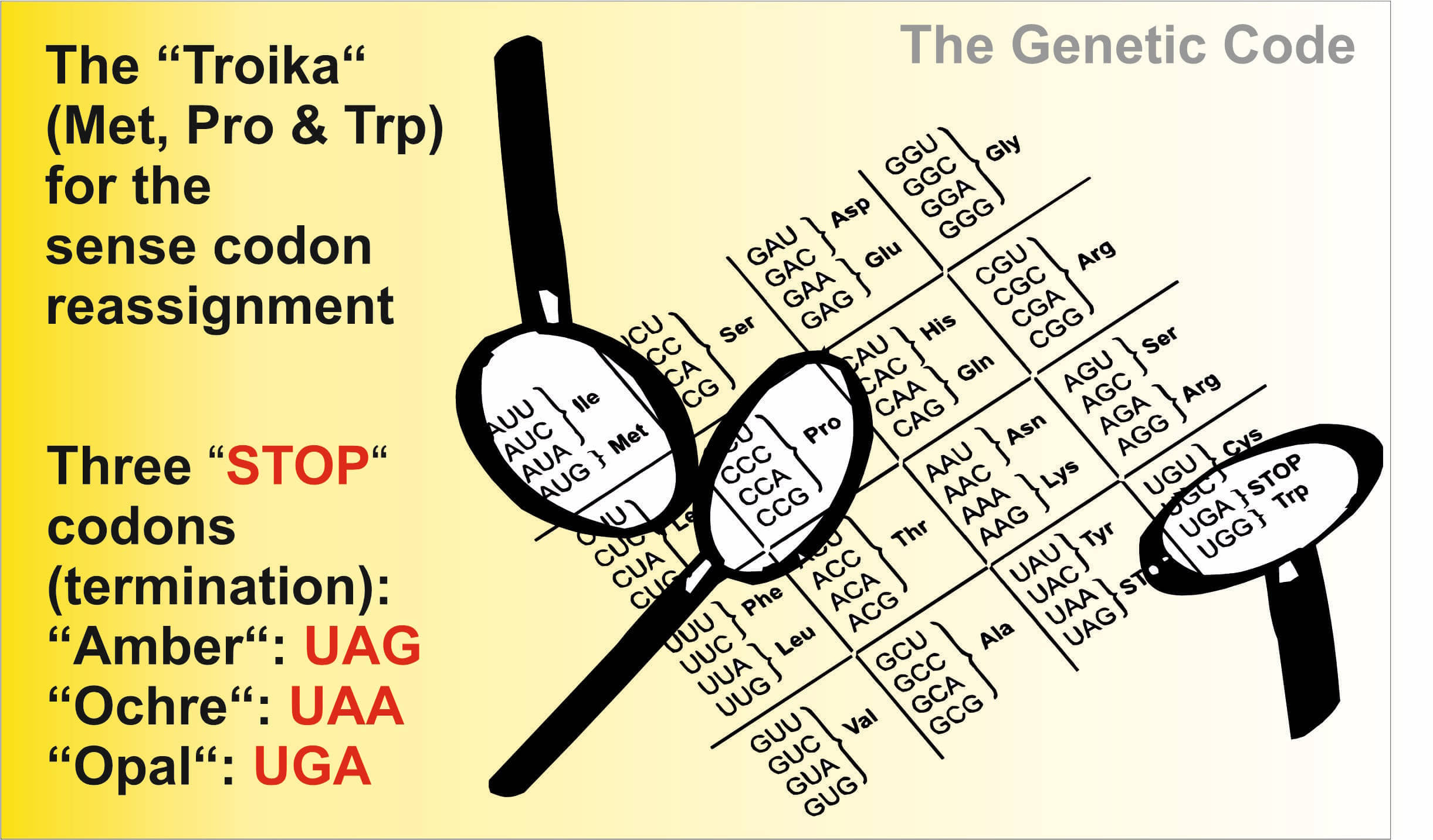
as targets for replacements (as well as the code amino acid repertoire expansions and reductions) with their analogs, surrogates, natural and synthetic counterparts with chemically changed side chains (Met, Trp) and protein backbone itself (Pro).
We perform mainly in vivo translations with various analogs or chemically similar structures of our "troika" in combination with their chemical or metabolic syntheses (transformations) using all available tools and approaches (including orthogonalized translation). We are also very interested to use their chemical analogs to delivery elements not used by nature (e.g. fluorine, tellurium and boron) into the biochemistry of life.
You will find more about proline, tryptophan and methionine in the Resources section, where all canonical members of the amino acid repertoire of the universal genetic code are presented in a comprehensible way.
Note that Proline (codons: CCU, CCA, CCG, and CCC) is considered to be a "primordial" amino acid and an integral part of the very early ‘GC code’. It has critical importance for protein folding and functions, in particular in repetitive oligopeptide motifs and processes that involve cis/trans isomerization.
On the other hand, both Met (AUG) and Trp (UGG) are considered as a "late addition" to the extant ‘GCAU code’ with special chemical functions in proteins. They are the least occurring amino acid in the proteome with low abundance in proteins (~1%) but highest metabolic synthesis cost (especially Trp) of all canonical amino acids.
Our main achievements with “Troika”:
Met
We were among the first groups who have used the Met analogues with selenium and tellurium-containing side chains for protein X-ray crystallography as well as to study and elucidate the mechanisms of folding and stability of proteins. For example, we made seminal contributions to understanding of the potential role of methionine oxidation in prion protein aggregation. In addition, we were among the first to use azide- and alkyne-containing Met analogues (originally developed by Dave Tirrell) for the "click" chemistry of proteins. Finally, we have pioneered in vivo metabolic engineering of Escherichia coli Met-biosynthetic pathways to generate diverse Met-like non-canonical amino acids that can be incorporated into proteins in situ. At present, we are interested in transforming the whole Met metabolism and translation by re-engineering methionine adenosyltransferase and Met-tRNA synthetases with sense codon reassignments.
Reading for more in-depth coverage of the subject
- Schipp, C. J., Ma, Y., Al-Shameri, A. Contestabile, R, Budisa, N. and DiSalvo, M.L. (2020) An Engineered Escherichia coli Strain with Synthetic Metabolism for In-cell Production of Translationally Active Methionine Derivatives. Chembiochem 21(24), 3525 – 3538; doi: 10.1002/cbic.202000257
- Nojoumi, S.; Ying, M.; Schwagerus, S.; Hackenberger, C.P.R.; Budisa, N. (2019) In-Cell Synthesis of Bioorthogonal Alkene Tag S-Allyl-Homocysteine and Its Coupling with Reprogrammed Translation. Int. J. Mol. Sci. 20 (9) e 2299; doi: 10.3390/ijms20092299
- Ma, Y., Biava, H., Contestabile, R., Budisa, N. & M.L. di Salvo (2014) Coupling Bioorthogonal Chemistries with Artificial Metabolism: Intracellular Biosynthesis of Azidohomoalanine and Its Incorporation into Recombinant Proteins. Molecules, 19(1), 1004-1022; doi: 10.3390/molecules19011004
Trp
In 2015, we reported the first successful completion of a long-term evolution experiment that resulted in full, proteome-wide substitution of virtually all 20,899 tryptophan residues with thienopyrrole-alanine in the genetic code of the bacterium Escherichia coli. These experiments were assisted with simple metabolic conversion of indole and indole-like analogs which we used to feed bacterial cultures. We are generally interested to transfer various physicochemical properties and special spectroscopic features (e.g. blue and golden fluorescence) of various indoles (and indole-like molecules) into the proteins of living cells. We also pioneered the use of Trp-like (and Tyr-like) analogs (often by including orthogonal translation) to create novel ribosomally synthetized peptide-drugs and new-to-nature biomaterials (e.g. photoactivatable mussel-based underwater adhesives). Currently, we are interested in combining evolutionary adaptations with the redesign of indole metabolism to produce bio-containing bacterial cells.
Reading for more in-depth coverage of the subject
- Hoesl, M. G., Oehm, S., Durkin, P., Darmon, E., Peil, L., Aerni, H.-R., Rappsilber, J., Rinehart, J., Leach, D., Söll, D. and Budisa, N. (2015) Chemical evolution of a bacterial proteome. Angew. Chem. Int. Ed. Engl., 54, 10030-10034; doi: 10.1002/anie.201502868
- Lepthien, S., Hoesl, M. G., Merkel, L., Budisa, N. (2008) Azatryptophans endow proteins with intrinsic blue fluorescence. Proc. Natl. Acad. Sci. USA. 105 (42), 16095-16100; doi: 10.1073/pnas.0802804105
- Pal, P. P. and Budisa, N. (2004) Designing novel spectral classes of proteins with tryptophan-expanded genetic code. Biol. Chem. 385(10), 893-904; doi: 10.1515/BC.2004.117
Pro
At the early (and current) stages of our research, we pioneered ribosomal in vivo and in vitro incorporations of Pro analogs which enabled us to discover the role of proline side-chain conformations (endo-exo isomerism) in protein translation and folding. Combined with peptide syntheses we are studying dynamics and stability of peptides and proteins especially by using fluorine-containing Pro analogs suitable for 19F NMR-spectroscopy.
Why do we so passionately like proline? It is a fundamentally different amino acid when compared to other members of the canonical amino acid repertoire. This makes Pro not only a unique target for protein research, but also a useful tool to question/ challenge our current concepts, wisdom and logic behind the amino acid repertoire establishment in evolution and the "frozen" code. Even more, the chemical diversification of Pro-structure may be the most promising starting point for attempts to create an alternative life structures from the first principles (and technologies derived thereof). This “parallel” life should be based on completely different (orthogonal) protein building blocks and related metabolites.
Reading for more in-depth coverage of the subject
- Kubyshkin, V., Davis, R., Budisa, N. (2021) Biochemistry of fluoroprolines: the prospect of making fluorine a Bioelement. Beilstein Journal of Organic Chemistry, 2021, 17, 439–460; doi: 10.3762/bjoc.17.40
- Larregola, M., Moore, S., Budisa, N. (2012). Congeneric bio-adhesive mussel foot proteins designed by modified prolines revealed a chiral bias in unnatural translation. Biochem. Biophys. Res. Commun. 421, 646-50; doi: 10.1016/j.bbrc.2012.04.031
- Moroder, L., Budisa, N. (2010) Synthetic biology of protein folding. ChemPhysChem 11(6), 1181-7; doi: 10.1002/cphc.201000035
Why code engineering?
In the extant living beings, the set of 20 canonical amino acids prescribed by the universal genetic code does not span all dimensions of chemical variability necessary for diverse cellular processes advanced (e.g. metazoan) cellular forms.
Keep always in mind: only a few proteins have a final covalent structure which is a simple accurate translation of mRNA. Thus, the release of proteins from a ribosome is usually not the last chemical step in their functional maturation to become e.g. structural components of the cell or to serve as functional biochemical machines.
The majority of the proteins reach maturity and converts into active forms, either co-translationally or by post-translational processing. Indeed, evolution invented these two strategies to increase amino acid side-chain inventory. First, a small fraction of proteins is co-translationally equipped with special proteinogenic amino acids such as selenocysteine (Sec) and pyrrolysine (Pyl) by reassignment of termination codons. The second and major class of chemical modifications that contribute to the protein structure/ function diversity are post-translational modifications (PTMs). These reactions are selectively and timely coordinated events performed by dedicated enzymes or enzymatic complexes, usually in a specialized cell compartment.
Why then code engineering? Indeed, we have PTMs that encompass all the dimensions of chemical variability necessary for different cellular processes and functions.
Nonetheless, there are at least three good reasons (all good things go by three) to use code engineering.
- PTM machinery is highly complex (compartmentalization!)
- It is difficult to achieve large quantities of homogeneous product
- The recognition features for specific PTM are normally easily destroyed in laboratory settings
It is extremely difficult to mimic nature's complex machines or processes such as the PTM. Therefore, we will require genetic code engineering as long as research in microfluidic devices and systems provides us with suitably miniaturized devices or micro-devices. For example, efficiently closed membrane or semipermeable orthogonal compartments such as vesicles might act as containers (or even protocells!) with encapsulated biochemical processes.
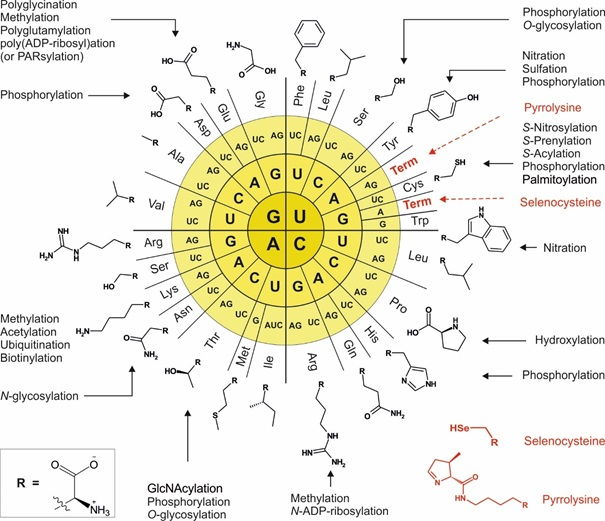
Reading for more in-depth coverage of the subject
- Kubyshkin, V. and Budisa, N. (2017) Synthetic alienation of microbial organisms by using genetic code engineering: why and how? Biotechnol. J., 12 (8), 1600097; doi: 10.1002/biot.201600097
- Hoesl, M. G., Budisa, N. (2012) Recent advances in genetic code engineering in Escherichia coli. Curr. Opin. Biotechnol. 23, 751–757; doi: 10.1016/j.copbio.2011.12.027
- Budisa, N. (2004) Prolegomena to future experimental efforts on genetic code engineering by expanding its amino acid repertoire. Angew. Chem. Int. Ed. Engl. 43(47), 6426-63; doi: 10.1002/anie.200300646
Academic application and potentials
Over the past 20 years, Ned teams have worked with various groups on joint projects. Many of these collaborations were very successful. Thereby, non-canonical amino acids proven to be valuable and useful tools for various research fields.
- Drug Design – code engineering as a route to diversify small bioactive molecules
- Biomaterials – new-to-nature congeneric peptides, proteins & complex scaffolds
- Bioorthogonal conjugations and chemistries – custom-made in vivo chemistries
- Structural Biology – noninvasive markers for 3D structures determination
- Fluorine biochemistry – fluorine as element of life and tool for NMR spectroscopy
- Spectroscopic probes in proteins – electron/proton pathways, signal transduction
- Photobiology – chromophore design (photophysics & optogenetics)
- Biophysics – protein folding, stability & energy dissipation pathways
- Biocatalysis – biocatalytic processes in cellular systems & enzymatic cascades
- Metabolic Engineering – reprograming intracellular amino acid syntheses
- Directed evolution – engineering & selection of enzymes and bacterial strains
Reading for more in-depth coverage of the subject
- Heidari-Karbalaei, H-R. and Budisa, N. (2020) Combating antimicrobial resistance with new-to-nature lanthipeptides created by genetic code expansion. Front Microbiol, 1:590522; doi: 10.3389/fmicb.2020.590522
- Agostini, F., Völler, J.-S., Koksch, B., Acevedo-Rocha, C. G., Kubyshkin, V. and Budisa, N. (2017) Biocatalysis with Unnatural Amino Acids: Enzymology Meets Xenobiology. Angew. Chem. Int. Ed. Engl., 56, 2-26; doi: 10.1002/anie.201610129
- Biava, H., Budisa, N. (2014) Evolution of fluorinated enzymes: An emerging trend for biocatalyst stabilization. Eng. Life Sci., 14, 340-351; doi: 10.1002/elsc.201300049
Xenobiology, Synthetic Biology and related Technologies
Latest at the end of the 20th century it become clear that engineering of e.g. different microbial strains for fermentative production is much more efficient than classical chemical engineering. For example, Danielli predicted that “the production of sequence-determined polymers rather than the random polymers…” will create a “degree of sophistication…. far beyond thought”. In 1971 he said: “I am convinced that so many people – even biologists – aren’t aware that biology is moving from an age of analysis into an age of synthesis” [New Scientist, 1971, pp. 124].
While Danielli in 1970th was a voice in the wilderness, nowadays is well established that the main goal of Synthetic Biology (SB) is the creation of biodiversity applicable for biotechnological needs. In contrast, Xenobiology (XB) aims to expand the framework of natural chemistries with the non-natural building blocks in living cells to accomplish artificial biodiversity.
The molecules, molecular complexes and processes along the flow of genetic information ("central dogma") are particularly attractive targets for such engineering. For example, the development of alternative nucleic acids (xenonucleic acids, XNAs) based upon new base pairs, sugars and backbones. Alternatively, estranging the genetic code from its current form via systematic introduction of non-canonical amino acids should enable the development of bio-containment mechanisms in synthetic cells potentially endowing them with a ‘genetic firewall’ i.e. orthogonality which prevents genetic information transfer to natural systems.
In general, XB is an attempt to produce non-natural molecules (xenobiotic materials) using chemically modified organisms that may be endowed with a genetic firewall. In this context, processed products derived from microbes (e.g. E. coli, B. subtilis, S. cerevisiae) are particularly attractive. Such microbes are generally very suitable ‘workhorses’ for redesigning, recoding, re-inventing natural processes within short timescales.
According to Phillipe Marliere both Xenobiology and Synthetic biology have following industrial goals:
- Produce chemicals, materials and energy vectors from renewable resources.
- Design, construct and evolve microbes with novel metabolic cores and coding pathways.
- Propagate synthetic eco-systems and food-chains.
- Diversify modes of element cycling and fixation (CHNOPS + Fluorine + Silicon + Boron)
- Promote evolutionary bifurcations, prevent genetic pollution (Biocontainment, Genetic firewall).
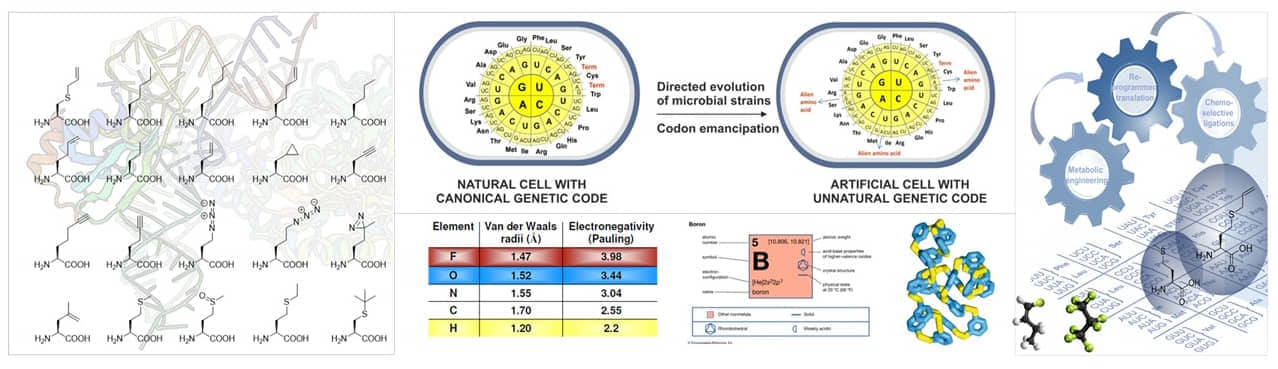
New to-nature xenobiotic materials of future will be elaborated through combination of bio-inspired modules that will greatly surpass existing technologies with potentially high value for the society as a whole.
Reading for more in-depth coverage of the subject
- Acevedo-Rocha, C. G., Budisa, N. (2011). On the Road towards Chemically Modified Organisms Endowed with a Genetic Firewall. Angew. Chem. Int. Ed. Engl. 50 (31), 6960–6962; doi: 10.1002/anie.201103010
- Budisa, N. (2014) Xenobiology, New-to-Nature Synthetic Cells and Genetic Firewall. Curr. Org. Chem., 18, 936-943; doi: 10.2174/138527281808140616154301; Online
- Acevedo-Rocha, C. and Budisa, N. (2016) Xenomicrobiology: a roadmap for genetic code engineering. Microb. Biotechnol., 9, 666-676; doi: 10.1111/1751-7915.12398