News
The logic of code expansion : “Essential Logic and Facts Behind the Expansion of the Genetic Code: A Critical Assessment”.
Related Article
Luu, H.T.L.; Karbalaei-Heidari, H.R.; Budisa, N. (2025) Essential Logic and Facts Behind the Expansion of the Genetic Code: A Critical Assessment. ChemCatChem, e00809. (doi: 10.1002/cctc.202500809)
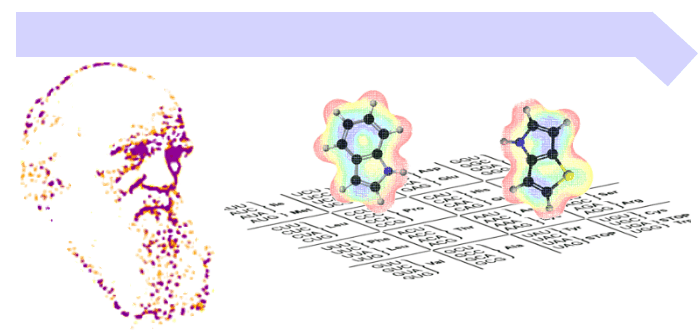
Life’s “operating system” is written in a universal genetic code. For billions of years it has been seen as frozen and unchangeable. Our article explains why this code looks the way it does - and how we are now finding ways to carefully expand it. This opens the door to designing new proteins, safer synthetic organisms, and future biotechnologies - but also raises deep questions about the very limits of life itself. We trace the chemical and evolutionary foundations of the genetic code, why life uses 20 amino acids, and how researchers are now working to expand this repertoire. The implications are vast - from new medicines to safe synthetic cells.
🔬 Is the “expanded genetic code” really an expansion? Efficiency, not tricks, will decide the future of genetic code expansion.
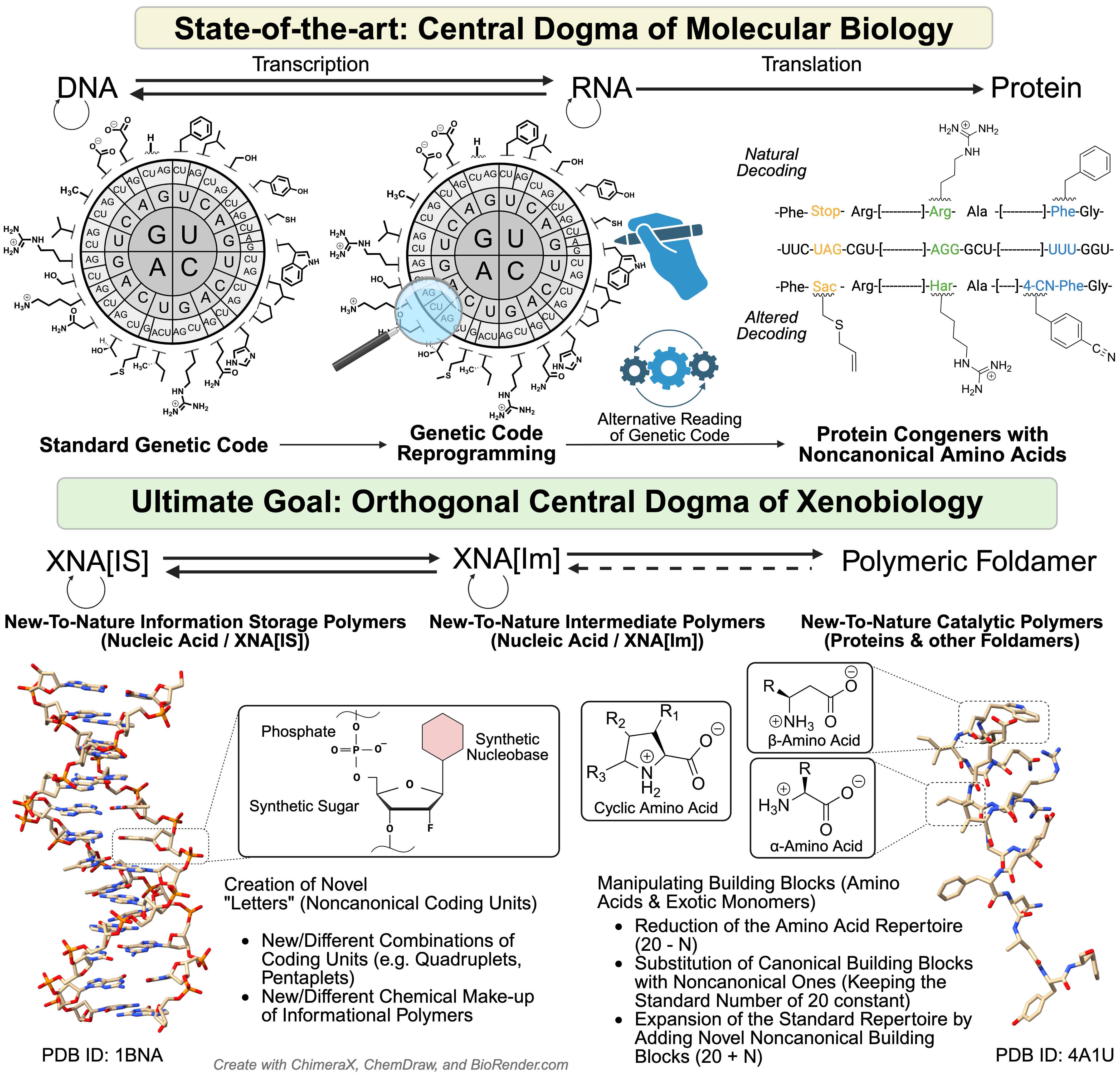
In our new paper (ChemCatChem), we argue that much of what is called “genetic code expansion” may actually be closer to tinkering with nature’s built-in recoding tricks (like stop-codon readthrough or frameshifting).
If we want to move from curiosity-driven experiments to industrial-scale biotechnology, efficiency is the key. Our results clearly show that as the number of in-frame stop codons increases, synthetic protein yield drops exponentially.
If this is the paradigm we present to the world as “ready for translational research,” we must ask: is this truly expansion — or just a fragile workaround?
🚀 Breaking the triplet barrier might be the key. Most genetic code “expansion” strategies recycle nature’s recoding tricks. But what if we re-engineered the architecture of the code itself?
In our new paper (ChemCatChem), we highlight a possible alternative: breaking triplet degeneracy — reshaping how codons are read, not just hijacked. Figure 7 (the Bohlke–Budisa experiment) illustrates how alternate genetic codes might be designed.
This is top-down synthetic biology. The other path — bottom-up design of entirely new, arbitrarily defined genetic codes — may be simpler in concept, but in practice we still don’t know how to do it (despite some bold claims in the literature).
This article has been impactful and recieved 14,000+ views over a 48 h period on X.
Maturing into a Revolutionary Tool : “Noncanonical Amino Acids” (Chem. Rev. virtual special issue)
Related Article
Budisa, N. (2025) Introduction: “Noncanonical Amino Acids”. Chem. Rev. 2025, 125, 1659−1662 (doi: 10.1021/acs.chemrev.5c00065)
Currently, almost 500 non-canonical amino acids (ncAAs) have been successfully incorporated into proteins by methods of expanding the genetic code, thereby expanding the repertoire of canonical amino acids (20) by an order of magnitude. These ncAAs, introduced primarily by anthropogenic innovation, incorporate novel chemical compounds and processes into the biochemistry of living systems, affecting everything from protein structures and functions to cellular genomes and proteomes. This progress is opening up new possibilities in synthetic biology, biotechnology, therapeutics, and materials science.
The special issue features 30 comprehensive reviews, covering nearly all major research areas and applications in the field. Jeffrey Wong, the pioneer of the Co-evolution Theory, outlines three distinct phases of genetic code development: from the RNA world, through metabolic integration, to a future era of synthetic life driven by noncanonical amino acids (ncAAs).

The incorporation of noncanonical amino acids (ncAAs) into proteins through ribosomal translation is redefining traditional biochemistry. This breakthrough enables the development of new therapeutic proteins, biocatalysts, and biocontainment strategies. Innovative techniques, such as residue-specific insertion using auxotrophic hosts, are proposed as more efficient alternatives to stop-codon readthrough, enhancing protein yields.
Proline analogues are gaining attention for their potential in drug development and protein engineering, while transfer RNAs (tRNAs) serve as key adaptors for ncAA incorporation, facilitating the creation of novel proteins and biomaterials through in vitro ribosomal synthesis. The “second genetic code” concept, introduced by Ellington, highlights the role of aminoacyl-tRNA synthetases (aaRSs) and ribosomal engineering in precisely integrating ncAAs into proteins. The special issue also delves into PylRS-based orthogonal translation systems, tRNA-based therapies for genetic disorders, cell-free protein synthesis (CFPS), bioorthogonal reactions, and the engineering challenges associated with mammalian systems.
While it is impossible to capture all the exciting developments in a brief overview, interested readers can explore Ned’s summary (doi: 10.1021/acs.chemrev.5c00065) and the entire special issue, which thoroughly covers current breakthroughs, concepts, and future directions. This collection is not only a rich resource on the cutting-edge research but probably also a historical document that highlights the evolving landscape of biological engineering in the 21st century.
Biosafe Escherichia coli: Genetically Firewalled Production Platforms with Built-In Orthogonal Components
Related Article
Karbalaei-Heidari, H.R., Budisa, N. (2024) Advanced and safe synthetic microbial chassis with orthogonal translation system integration. ACS Synthetic Biology 13 (9), 2992-3002 (doi: 10.1021/acssynbio.4c00437)
Synthetic life - whether created from inorganic matter or by chemically altering existing life forms - is not only one of the last sacrosanct frontiers in academic biosciences but also a potential breakthrough technology with revolutionary opportunities. Therefore, global research teams are advancing Synthetic Biology by engineering organisms to produce valuable substances, paving the way toward synthetic life.

Reprogramming protein translation with noncanonical amino acids (ncAAs) requires (a) metabolic compatibility, (b) translational orthogonality, and (c) high-fidelity ribosomal synthesis in living cells. Orthogonality ensures that directed evolution of aminoacyl-tRNA synthetases (aaRSs) yields robust enzymes selective for their cognate tRNA and ncAAs amidst endogenous tRNAs, aaRSs, metabolic intermediates, and 20 canonical amino acids. Our earlier work generated orthogonal variants with high activity and specificity for various ncAAs tolerated in the cytosol.
In vivo incorporation of isosteric amino acid analogs began over five decades ago. By the early 2000s, diverse analogs became vital tools for engineering proteins in auxotrophic bacteria. Both orthogonal translation and auxotrophy-based methods rely on plasmid-directed gene expression and optimized fermentation for residue-specific substitutions, but plasmid-based systems hamper breakthroughs in genetic code expansion and metabolic orthogonalization.
We addressed these constraints by engineering plasmid-free Escherichia coli chassis, providing robust synthetic cells to produce new-to-nature proteins and polymers with inherent biosafety. These genetic firewall-enabled cells integrate extended genetic codes into their genomes, incorporating over 300 ncAAs. Our work, employed these cells to synthesize fluorine-containing bioglues for wound healing and tissue engineering, demonstrating their stability and safety.
Towards a Synthetic Life Without Stress: Evolving Cells with 'Alien' Genetic Codes
Related Article
Tolle, I., Oehm, S., Hoesl, M.G., Treiber-Kleinke, C., et al. & Rappsilber, J. Ignatova, Z., Gerstein, A.C. Budisa, N. (2023) Evolving a mitigation of the stress response pathway to change the basic chemistry of life. Front. Synth. Biol. 1:1248065 (doi: 10.3389/fsybi.2023.1248065).
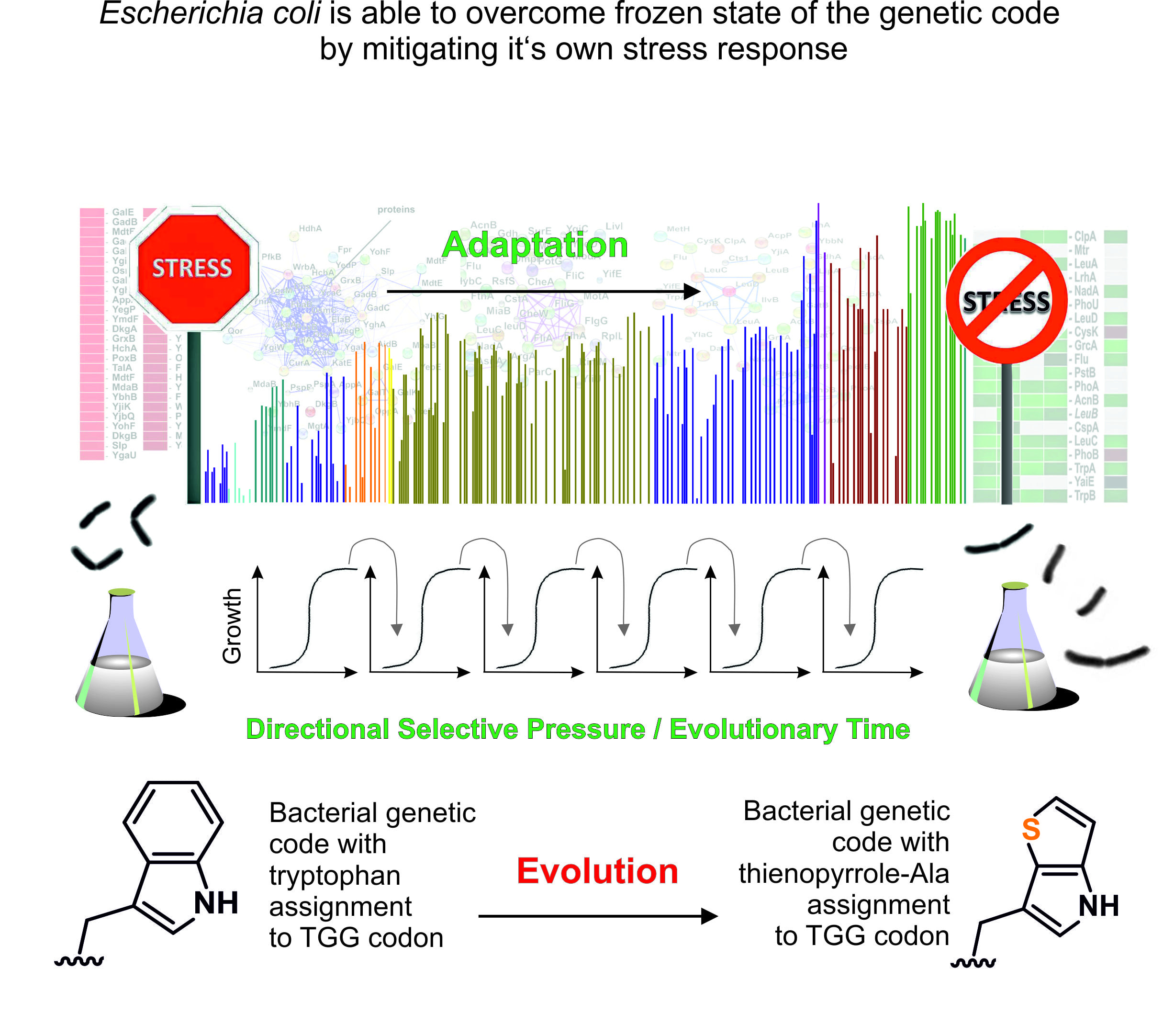
In 2015, we made made a groundbreaking achievement by evolving Escherichia coli to replace all tryptophan (Trp) residues in its proteome with the noncanonical amino acid (ncAA) analog Thienophenyl-alanine ([3,2]Tpa) using ALE (Hoesl et al., Angewandte Chemie, 2015). This was the first experiment of its kind to provide clear analytical proof that all Trp residues in the E. coli proteome were quantitatively replaced by an analog. Recent work by our team revealed that the gradual adaptation of E. coli to process and incorporate [3,2]Tpa was facilitated by mutations in genes involved in key physiological processes, particularly the general stress response. This response helps bacteria adapt to fluctuations in nutrients, temperature, pH, and osmolarity. The key discovery is that the conserved genetic code can be experimentally altered by addressing biological constraints, such as metabolic regulatory networks that maintain or "freeze" the code. This insight is crucial for ALE with synthetic cells and "alienated" genetic codes, providing a conceptual framework for the laboratory-driven reassignment of codons in the ‘universal’ genetic code. The complete progress of this work is reviewed by in Frontiers in Synthetic Biology (https://doi.org/10.3389/fsybi.2024.1433522).
Cutting-Edge Molecular Probes: Exploring Hydrogen Bonds, Vibrational Energy, and Electric Fields
Related Article
von Sass, G.J., Blain‐Hartung, M., Baumann, T., Forest, K.T., Hildebrandt, P., Budisa, N. (2023) Orthogonal translation with 5‐cyanotryptophan - infrared probe for local structural information, electrostatics, and hydrogen bonding. Protein Sci. 32(7):e4705 (doi: 10.1002/pro.4705).
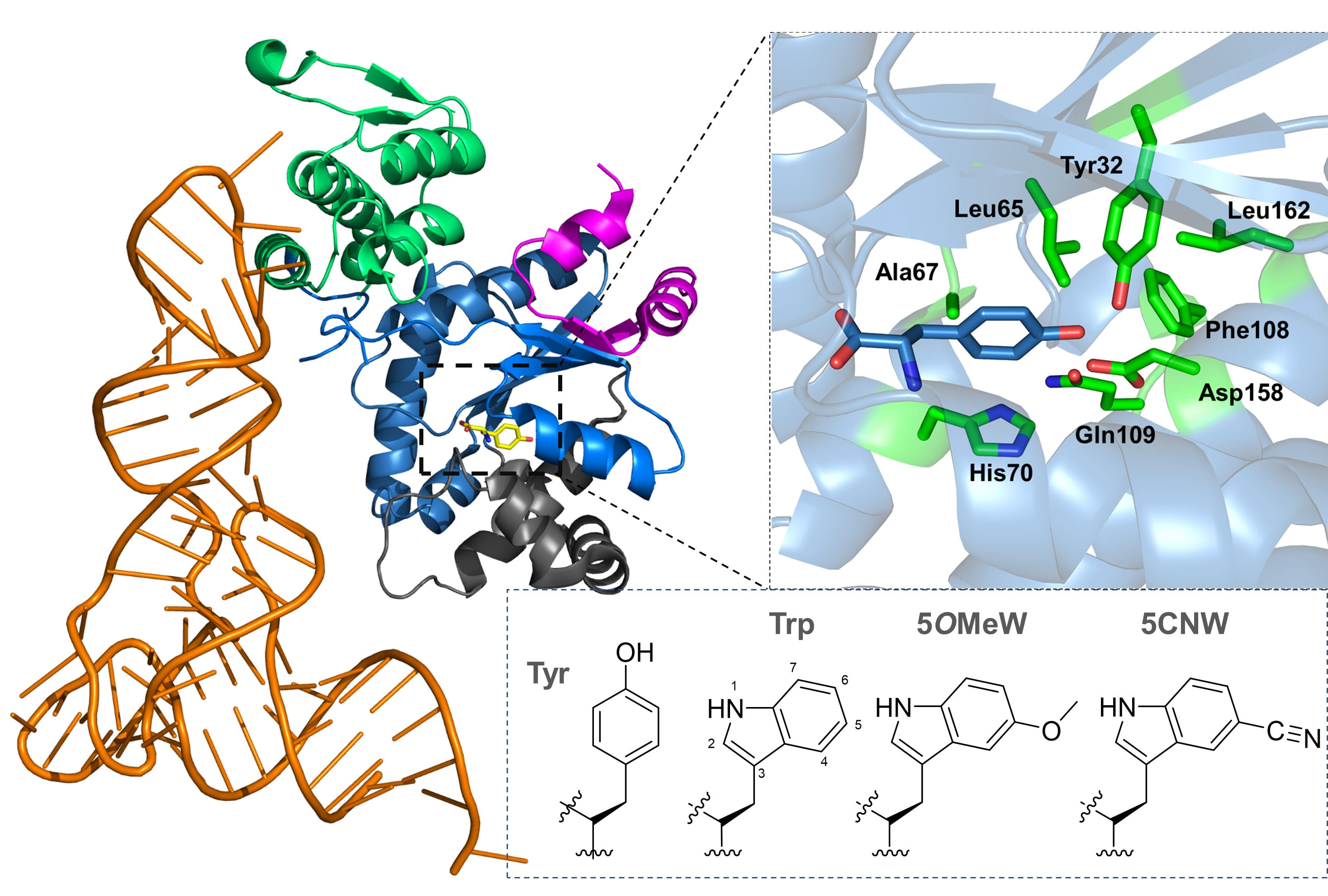
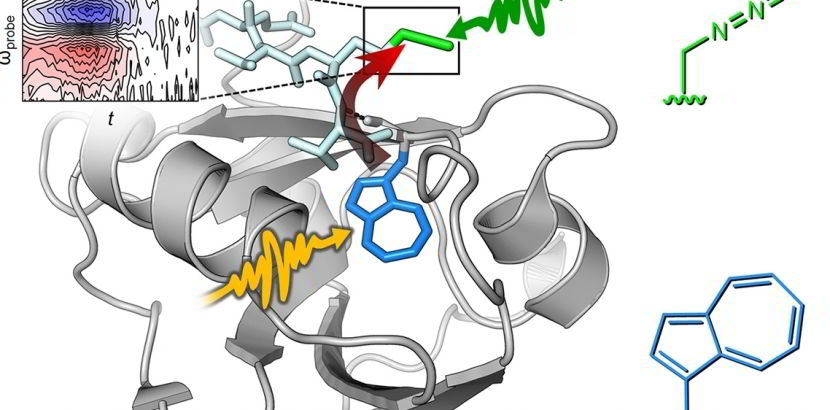
Nitrile-containing tryptophan analogs, such as 5-cyanotryptophan (5CNW), are valuable probes for studying local electrostatics and hydrogen bonding in rigid and dynamic environments. The nitrile (CN) group in 5CNW provides non-invasive labeling in proteins, offering detailed insights into local electrostatics and hydrogen bonding through IR spectroscopy. This versatile probe supports both static and dynamic measurements.
Tryptophan (Trp) itself is a powerful biophysical probe due to its unique fluorescence properties, sensitivity to microenvironments, and ability to interact with molecules of varying polarity and ionization states. However, its intrinsic indole side chain can be limiting for probing phenomena like local electric fields or vibrational energy transfer. Chemically diversifying the indole side chain, such as substituting Trp with 5CNW, greatly enhances its utility. 5CNW, an electron-withdrawing Trp surrogate, exhibits significant changes in quantum yield and fluorescence lifetime, making it an excellent tool for parameterizing local interactions via IR spectroscopy.
We developed an orthogonal translation system to incorporate 5CNW into a photochromic photoreceptor protein, enabling non-invasive monitoring of local electrostatics through the Vibrational Stark Effect. Leveraging the evolutionary similarity between tryptophanyl-tRNA synthetase (TrpRS) and tyrosyl-tRNA synthetase (TyrRS), we used MjTyrRS as a starting point to engineer recognition of Trp analogs, given Trp's evolutionary divergence from Tyr as a late addition to the genetic code.
Our work has demonstrated the utility of ncAAs as valuable probes for structural biology and biophysics. Initially applied in X-ray crystallography, their use later expanded to biophysical studies. These probes have uncovered key phenomena, including proline puckering's role in protein folding, the development of click chemistry and bioorthogonal conjugation techniques, and the non-invasive detection of local electric fields within protein interiors (in collaboration with Dr. P. Hildebrandt and Dr. T. Friedrich at TU Berlin). Additionally, ncAAs have been essential in tracing Vibration Energy Transfer (VET) in complex protein structures (in collaboration with Dr. J. Bredenbeck at Goethe University, Frankfurt). These findings have been highlighted in numerous publications over the past decade.
Keeping Proteins in Check: Using Orthogonal Translation to Tackle Stress-Induced Modifications Linked to Aging and Disease
Related Articles
Sun, H., Jia, H., Kendall, O., Dragelj, J., Kubyshkin, V., Baumann, T., Mroginski, A.A., Schwille, P., Budisa, N.* (2022) Halogenation of tyrosine perturbs large-scale protein self-organization. Nat. Commun.13, e4843 (doi: 10.1038/s41467-022-32535-2). [PI; 50%]
Koch, N. G., Baumann, T., Nickling, J. H., Dziegielewski, A., & Budisa, N.* (2022). Engineered bacterial host for genetic encoding of physiologically stable protein nitration. Front. Mol. Biosci. 9, e992748 (doi: 10.3389/fmolb.2022.992748). [PI; 50%]
Proteins are essential for life, performing diverse functions such as catalysis, structural support, and signaling. They are synthesized from 20 canonical amino acids (cAAs), which can undergo post-translational modifications (PTMs) that expand protein functionality. PTMs, both enzymatic and non-enzymatic, involve chemical changes like glycation, nitrosylation, oxidation, acetylation, and halogenation. These modifications often alter tertiary structures, destabilize proteins, or lead to aggregation, impacting protein quality and cellular functions. Oxidative halogenation, a pathological modification, contributes to aging, cancer, and other health issues. Tyrosine is particularly susceptible, with hypochlorous acid from immune cells being a major source of 3-chlorotyrosine formation. Additionally, environmental halogenated compounds raise concerns about acquired halogenation.
Genetic incorporation of noncanonical amino acids (ncAAs) provides a powerful approach to study PTMs. Co-translational insertion of oxidized or halogenated ncAAs enables precise analysis of their structural and functional effects. For example, nitrotyrosine has been incorporated into mitochondrial superoxide dismutase to investigate oxidative damage, while modifications like acetylation and methylation have revealed their roles in nucleosome assembly and transcription.
Our work introduces two breakthroughs: (1) a redesigned tyrosyl-tRNA synthetase (MjTyrRS) for producing stable photo-caged polypeptides for biomaterials and antibiotics, and (2) a platform for site-specific incorporation of halogenated tyrosine analogues in E. coli. We eliminated endogenous nitroreductase activity via genome engineering to enhance spatiotemporal control of caging. These advances enable precise studies of oxidative damage, offering insights into aging and cancer, and paving the way for improved diagnostics and therapies.

Solubility Matters: How a Tiny Tag Turbocharges Genetic Code Expansion
Related Article
Koch, N.G. Baumann, T., Budisa, N. (2021) Efficient unnatural protein production by pyrrolysyl-tRNA synthetase with genetically fused solubility tags. Frontiers in Bioengineering and Biotechnology, e1247 (doi: 10.3389/fbioe.2021.807438).

The solubility of pyrrolysyl-tRNA synthetase (PylRS) expressed in Escherichia coli is limited due to its aggregation tendency and incompatibility with bacterial folding systems. Issues such as misfolding, inclusion body formation, and inefficient folding often arise during high-level expression. These challenges reduce ribosomal ncAA incorporation efficiency and enzyme activity, ultimately leading to low yields of target proteins.
Several strategies have been employed to address these challenges, including codon optimization, adjustments to expression conditions (e.g., reduced temperature or IPTG concentration), co-expression of chaperones like GroEL/GroES, and protein engineering. Fusion with solubility tags such as MBP, GST, or SmbP significantly improves solubility and stability. The SmbP-tag, derived from a small metal-binding protein in Nitrosomonas europaea, effectively enhances the solubility of aggregation-prone proteins in E. coli.
We demonstrated that N-terminal fusion of SmbP increases PylRS solubility and boosts orthogonal translation efficiency by 200–540% for engineered variants and 245% for the wild-type enzyme. This strategy enhances the incorporation of unnatural amino acids into proteins, enabling efficient site-specific labeling and triple labeling capabilities.
Protein solubility is essential for maintaining enzymatic functionality and ensuring effective ncAA incorporation. By combining solubility tag fusion with optimized expression conditions, we have successfully overcome solubility bottlenecks in PylRS, providing a reliable tool for synthetic biology and genetic code expansion applications.
Small Tags - Big Impact: Advancing Genetic Code Engineering with Small-Chain Noncanonical Amino Acids
Related Article
Koch, N.G., Goettig, P., Rappsilber, J. & Budisa, N. (2021). Engineering pyrrolysyl-tRNA synthetase for the incorporation of non-canonical amino acids with smaller side chains. Int J Mol Sci 22 (20), e11194 (doi: 10.3390/ijms222011194).
The majority of engineered aminoacyl-tRNA synthetase (aaRS) variants are derived from Methanosarcina mazei/barkeri pyrrolysyl-tRNA synthetases (MmPylRS/MbPylRS) or Methanocaldococcus jannaschii tyrosyl-tRNA synthetase (MjTyrRS). Their archaeal origin and distant phylogeny ensure orthogonality in bacterial cells, enabling the seamless implementation of orthogonal translation systems (OTSs). PylRS/tRNAPyl^\text{Pyl}Pyl pairs from Methanosarcina species are particularly valuable for genetic code expansion (GCE) due to their orthogonal activity in both bacterial and eukaryotic systems.
The wild-type PylRS, which activates the native substrate pyrrolysine, can also incorporate structurally similar ncAAs. This remarkable substrate promiscuity arises from nonspecific hydrophobic interactions within its large binding pocket, enabling the recognition of a wide range of ncAAs with minimal mutations (2–4). However, most ncAAs compatible with the PylRS system are characterized by flexible, long-chained, and bulky pyrrolysine analogs or shorter yet bulky aromatic substrates, such as phenylalanine, tryptophan, and histidine analogs.
Our study marks a significant breakthrough in the field by utilizing SacRS, a PylRS variant engineered to incorporate S-allyl-cysteine (Sac), to develop new MbPylRS variants capable of incorporating small sidechain ncAAs. This advancement allows PylRS-based OTSs to incorporate an unprecedented diversity of side chains into proteins, including small, large, bulky, aromatic, polar, hydrophobic, and bioorthogonal functionalities, as well as rare elements like fluorine and boron.
These ncAAs, categorized into five classes (see accompanying figure), can also include substrates with azido, fluoro, cyano, and nitroso groups, which are highly valuable for FTIR, NMR, crosslinking, and spin labeling. This approach addresses unmet needs, such as designing ncAA analogs for Glu and Asp or mimicking post-translational modifications, significantly broadening the scope of GCE and transforming applications in protein engineering.

Fluoroprolines: A Twist of Fluorine for Next-Level Protein Engineering
Related Article
Kubyshkin, V., Davis, R., Budisa, N. (2021) Biochemistry of fluoroprolines: the prospect of making fluorine a Bioelement. Beilstein Journal of Organic Chemistry, 2021, 17, 439–460 (doi: 10.3762/bjoc.17.40).

Proline (Pro) stands out within the canonical amino acid repertoire due to its unique N-alkylamino structure with a pyrrolidine ring. This distinctive feature affects proline's reactivity and the trans-cis isomerization of the peptidyl-prolyl amide bond, significantly influencing protein structure and function. The trans and cis isomers of the peptidyl-proline bond exhibit varying stabilities depending on the exo and endo pucker conformations of the pyrrolidine ring. These conformations can be systematically stabilized or destabilized by specific 4-substituents, enabling control over proline's conformational preferences.
The incorporation of synthetic C-4 fluoroprolines, such as (4R)-fluoroproline (Flp), (4S)-fluoroproline (flp), and 4,4-difluoroproline (dFP), into peptides and proteins has revealed their profound effects on ring puckering, kinetics, structure, and stability. Studies over the last two decades demonstrate that replacing proline with fluoroprolines enhances protein expression speed and yield while improving thermodynamic and kinetic folding profiles.
This review summarizes the biochemical role of proline, the physicochemical properties of fluoroprolines, and their applications as proline replacements in protein production and folding studies. Fluoroprolines have emerged as valuable tools in biotechnology, with potential for proteome-wide proline-to-fluoroproline substitutions. As a future prospect, we propose leveraging laboratory evolution methods to introduce fluoroprolines into living cells, enabling the creation of synthetic cells with artificial biodiversity. This approach could establish fluorine as a novel bioelement and unlock new frontiers in synthetic biology.
Meet an Alien: Xenobiology is coming of age!
Xenobiology has now matured intellectually and experimentally as a scientific discipline. It is expected to provide us with a good starting point and platform for current and future attempts to fundamentally remake life or even create life forms from scratch!
With the publicly available Special Collection Xenobiology 2020, we will get much clearer ideas and inspiration on how to completely change the chemical implementation of life (both top-down and bottom-up), which means to synthetically (re-)design every single element of life (see figure below).
The diversity in nature usually leads to the creation of species that are "reproductively isolated" (classic definition of species). In Xenobiology however, engineered diversification (top-down) and de novo (from scratch) synthesis (bottom-up) leads to the creation of contained parallel biological worlds in genetic isolation.
Since coding is an essential feature of life, we expect that real artificial, synthetic (alien) life will be equipped with novel genetic codes! You can read more about this in our editorial for Special Collection of ChemBioChem Xenobiology 2020.
However, it could be misleading when talking about “creating aliens”. Extraterrestrials might already exist somewhere in space (or maybe on Earth, hidden from us). When we talk about aliens in everyday life, we always have extraterrestrials in mind. However, the ‘aliens’ of Xenobiology are no extraterrestrials. They are our creations!
Our recent proposal of the “Alanine World” might be helpful (to some extent) in this regard. When we talk about our world as the Alanine-World, it helps us to clearly demarcate it from ‘XB aliens’ as well as possible aliens of Extraterrestrial Biology and Astrobiology. But who guarantees that extraterrestrial or ‘hidden’ aliens are not based on the Alanine World as well? Maybe we are living in the best of all possible worlds. Who knows?
This is a good point to stop and go back to the boring mainstream science and do something in the laboratory…
Genetic code engineering meets nanotechnology
Selective targeting of cellular receptors is the main strategy to block the influenza virus, which is adapted by viruses and other parasites to infect the host cell. This event could be blocked or significantly disrupted by the development of specific binders that act on either cellular receptors or by blocking pathogen recognition proteins. This is exactly what we have been doing in cooperation with various Berlin groups over the past six years.
For further information, take a look at Science Daily and University of Manitoba press release.
Original paper: Nature Nanotechnology, 2020; DOI: 10.1038/s41565-020-0660-2
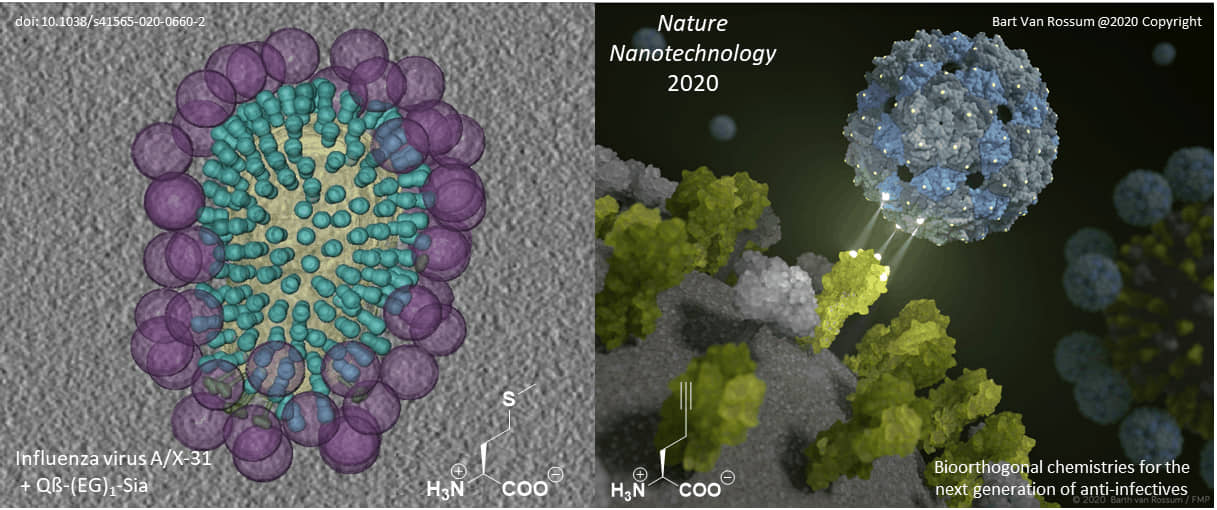
Proteins with well-characterized three-dimensional structures are ideal frameworks for nanotechnology. Their defined and rigid three-dimensional structure enables the positioning of ligands with an almost perfectly defined geometry. Most importantly, our genetic code engineering enables us to endow target protein scaffolds and their assemblies (such as viral capsids) with bioorthogonal chemical functionalities provided by non-canonical amino acids (ncAAs).
...or why did nature choose the genetic code with "only 20” canonical amino acids for ribosomal protein synthesis?
There seem to be several explanations for why nature chose 20 amino acids (as basic building blocks - monomers - for proteins) within the (almost) universal genetic code. Our contribution is a model of the "Alanine World", which is proposed from the viewpoint of peptide and protein chemistry. The core of our considerations is the choice of secondary structural elements for building up the protein scaffolds. This also determines the choice of amino acid monomers. Dominant secondary structures in life as we know it are α-helices and β-sheets, which are mainly made up of alanine derivatives (from a chemical point of view). That is why Vladimir coined the term "Alanine World". In this model, the selection of monomers (amino acids) for such secondary structural elements is rather limited. For example, chemical propensities for α-helix or β-sheet determine exactly which side chains of alanine derivatives are suitable and which are not.
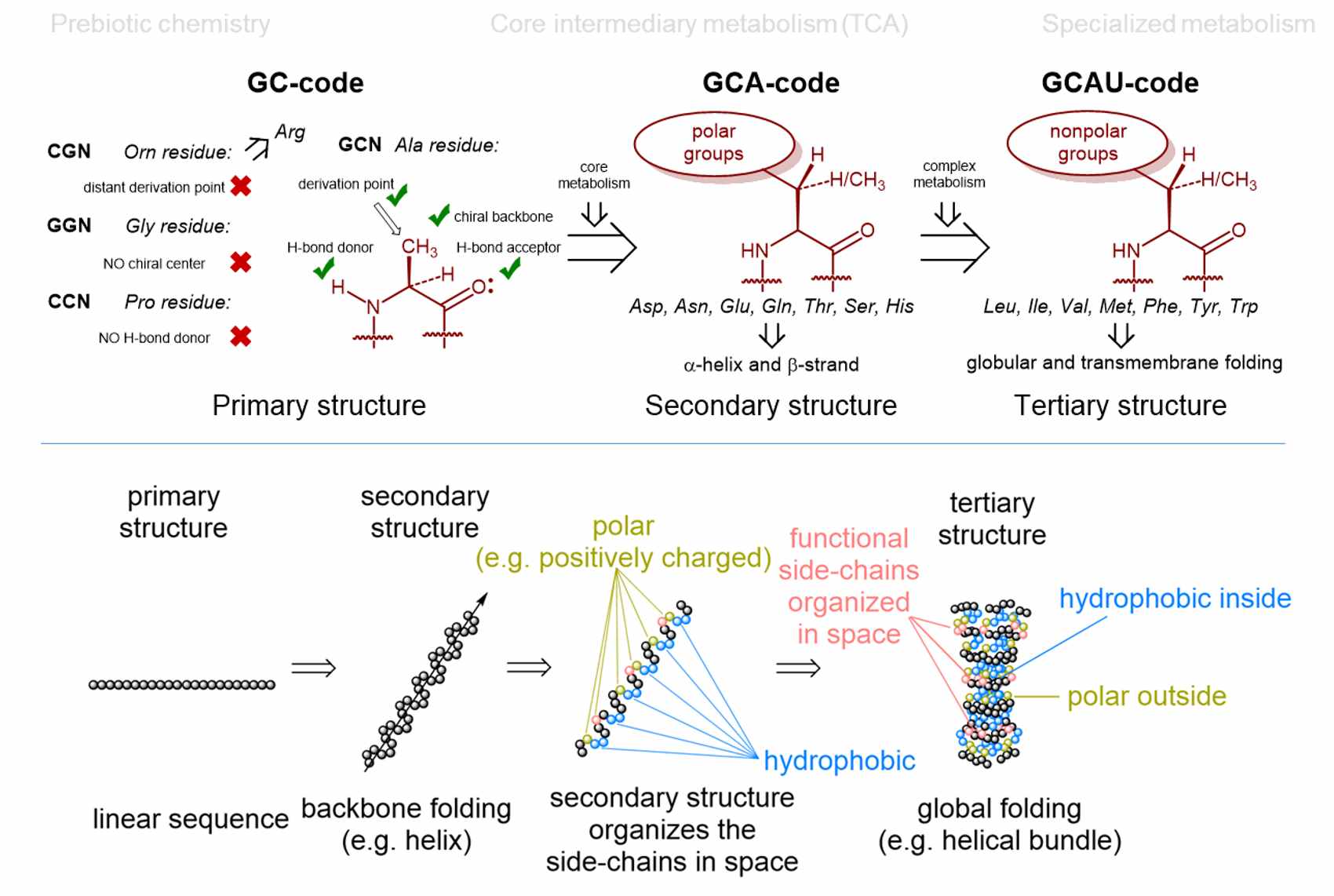
See our current papers for further details and comprehensive explanations as well as possible biotechnological relevance and impact.
Curiously, the numbers 3, 4, 64 and 20 in the flow of genetic information also seem to be magically attractive to quantum physicists. These numbers are obtained using strict quantum mechanical algorithms for the flow of genetic information (DNA -> RNA -> protein). The result is 4 letters (AUGC), 64 triplets and 20 monomers (amino acids) as optimal numbers.
That said, different scientific approaches allow the conclusion that the structure of the genetic code is deterministic. Does this mean that the existing living world is "the best of all possible" worlds? If so, the question arises whether it is worth at all to create an alternative to extant life?
Computational design takes over!
Computer-aided design provided an enzyme that surpassed all related enzymes generated by classical engineering methods.
Computational design and machine learning in the process of evolving highly active artificial enzymes is becoming an indispensable tool and is likely to completely replace classical approaches in the near future.
The context in which the group developed a novel, highly efficient orthogonal tyrosyl-tRNA synthetase (MjTyrRS), outperforming its natural ancestor enzyme through directed evolution and machine learning-guided design of focused enzyme libraries, was elaborated in Ned's 2021 talk to the Argentine Society of Organic Chemistry at Córdoba University.
In vivo Azulenyl-alanine incorporation into proteins by directed evolution of orthogonal enzymes
 For more information, see the preliminary conference report from the OPTICA Publishing Group, which announced this breakthrough in the field.
For more information, see the preliminary conference report from the OPTICA Publishing Group, which announced this breakthrough in the field. Excellent location in the land of ideas


Our project Xenoglue has recieved a award for its unique apporach and great potential. The price was awarded from the initiative "Deutschland-Land der Ideen" (Germany-land of ideas).
We are moving to Canada
The head of the group, Nediljko Budisa, has accepted a position at the University of Manitoba, located in Winnipeg, province Manitoba, Canada. We will officially move there starting from the 01.10.2018.


Prize for Xenoglue Team!

The technology of Xenoglue, which originates from our group, won the Science4Life start-up competition. Two represantatives of the Xenoglue-team, Christian Schipp and Tobias Schneider, took the award on the 13.03.2018 and the price money of 1000€. Before that they took part in a two day workshop about startup management. Xenoglue strives to develop a bio degradable glue, which is based on the natural example of the mussel and is intended for usage in wound healing. Scientist have studied the subject of the mussels adhesion mechanism for decades. (See news from 01.08.2017)
BIOMOD 2017: Gold winning MultiBrane team back from San Francisco!

The team MultiBrane of the tu project "iGEM Synthetic Biology" 2017 belongs to the big winners of this year's BIOMOD competition. The research objective was to create a multifunctional membrane for waste water treatment with a particular focus on microplastics & micropollutant removal. The results were very compelling and achieved high visibility in San Francisco, where the BIOMOD jamboree took place at UCSF University. Especially in three categories, MultiBranes convinced the international judges and received a gold medal for their project work:
> 3rd place in the best project website and
> 3rd place in the best video category
We would therefore like to express our sincere thanks to Saba Nojoumi and Franz-Josef Schmitt for their dedicated mentoring and support respectively. We also thank Hannah Aring and Nikolaj Koch for their work within the scope of tu projects. With this in mind, we congratulate again and wish all the best to all team members and look forward to the next exciting BIOMOD and iGEM projects in the future.
The work of the “MultiBrane - BiIOMOD Berlin 2017” team is summarized onYouTube.
Towards biological underwater superglue!

Regenerative medicine urgently needs biocompatible adhesives suitable for therapeutic use in the treatment of small bone fractures. Such an adhesive should allow rapid bonding of the bone fragments without the need for the attachment of plates and screws. The use of biological glue from marine mussels (so-called mussel-adhesive proteins, MAPs) - as potential environmentally-friendly bio-inspired adhesives - could be a solution to this problem. In nature, mussel secrets adhesive proteins and efficiently adheres to stone and other inorganic surfaces and even to man-made products such as metal and plastic (e.g. Teflon) materials. The secret of these bio-glues is the presence of catechol groups in the side chain of the non-proteinogenic amino acid L-dopa (L-3,4-dihydroxyphenylalanine) which is produced post-translationally by tyrosine hydroxylation and is capable of surface adhesion.
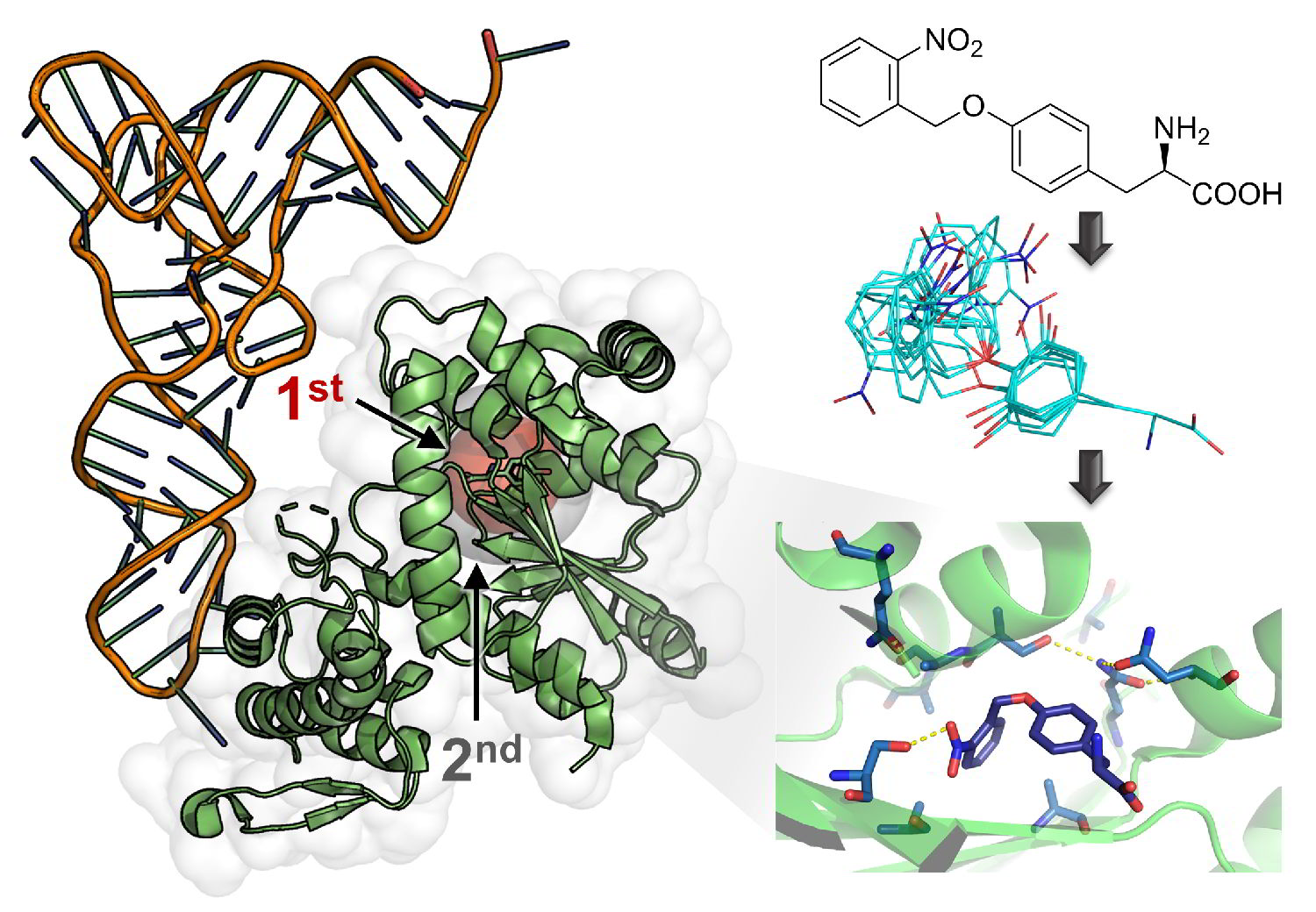
However, isolation of such bio-glues from natural sources is expensive, inefficient, and it is not possible to produce large amounts of a homogeneous product by imitating post-translational modification machineries. On the other hand, current synthetic chemical or biotechnological synthesis pathways are not efficient. For these reasons, we have endeavored to solve this problem by applying an orthogonal amino acid translation to develop a new concept to produce DOPA-rich underwater adhesive proteins from marine mussels by genetic code expansion.
By using computational design and genetic selection methods, a novel Methanocaldococcus jannaschii tyrosyl-tRNA synthetase-based enzyme was engineered. It activates the photocaged DOPA derivative ortho-nitrobenzyl-Dopa (ONB-DOPA) for incorporation into proteins in response to amber stop codons through orthogonal translation. In this way, mussel proteins are equipped with ONB-DOPA at multiple sites, which introduces spatiotemporal control over their adhesive properties. Exposure to UV light triggers cleavage of the ONB photocage, liberating the adhesive catechol side chain of DOPA. This strategy provides new ways for producing DOPA-based wet adhesives for application in industry and biomedicine with the potential to revolutionize bone surgery and wound healing.
Public resonance:Synthetic alienation of microbes - "thawing" of the "frozen" code
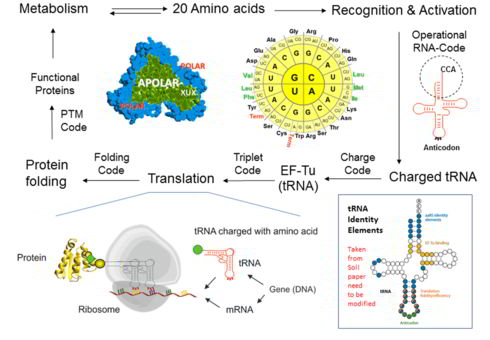
It is assumed that the genetic code experienced a transition from an ambiguous to a well-defined ("frozen") code, as we know it. During this evolution, the protein translation, which first operated with few amino acids and produced the statistical proteins, developed into a highly accurate machine capable of producing very specific proteins, based on the 20 canonical amino acids. Since the genetic code operates well in many ways (e.g. error minimization, protein folding, and metabolic integration), the fundamental question is, what are the barriers to attempts to change it experimentally? How can they be overcome? We have tried to find an answer and written an essay that is now published in the Biotechnology Journal (link) with a model that has allowed us to provide "entry points" for noncanonical amino acids in the ribosome protein synthesis cycle as shown above.