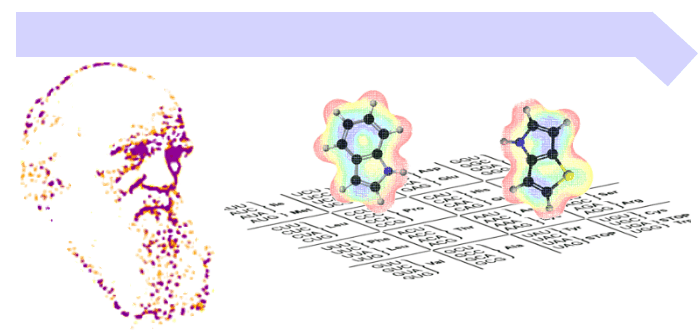
Publications
The group’s publication record and research funding are strong, with the PI ranked among the top five Chemistry researchers at the University of Manitoba.* International patent applications were filed (2021–2024), and IP ownership for a groundbreaking 2018 patent on wet bioadhesives has been granted in the USA, Canada, and most recently, China.
Lists with detailed metrics are available on:* see here