Running Projects
A Universal Synthetic Biosafe Microbial Chassis and Cell Factories

We employ genome editing to build robust, antibiotic-free biotechnological platforms that produce a diverse range of medically (e.g. lantibiotic-based) and industrially (different synthetic enzymes) important biopolymers (bio-contained bacterial cell factories). These next-generation whole-cell catalysts can be equipped with bioorthogonal functions for plastic degradation, sensing, remediation, genetically encoded click chemistry etc. opening new horizons in sustainable manufacturing and healthcare applications. These attributes collectively provide a robust, controllable “chassis” for building and testing new biological functions in a safe and universal way.
Reading for more in-depth coverage of the subject
- Heidari-Karbalaei, H-R. and Budisa, N. (2024) Advanced and Safe Synthetic Microbial Chassis with Orthogonal Translation System Integration. ACS Synth Biol. 13(9), 2992–3002; doi: 10.1021/acssynbio.4c00437
- Romero-Orejon, K., Karbalaei-Heidari, H.R., Budisa, N., Levin, D.B. (2025) Antibiotic-free whole-cell biocatalytic fermentation: Escherichia coli with surface-displayed PETases for sustainable plastic degradation. BioRxiv. doi: 10.1101/2024.11.20.624590
Expanding the Genetic Code by Directed Enzyme Evolution
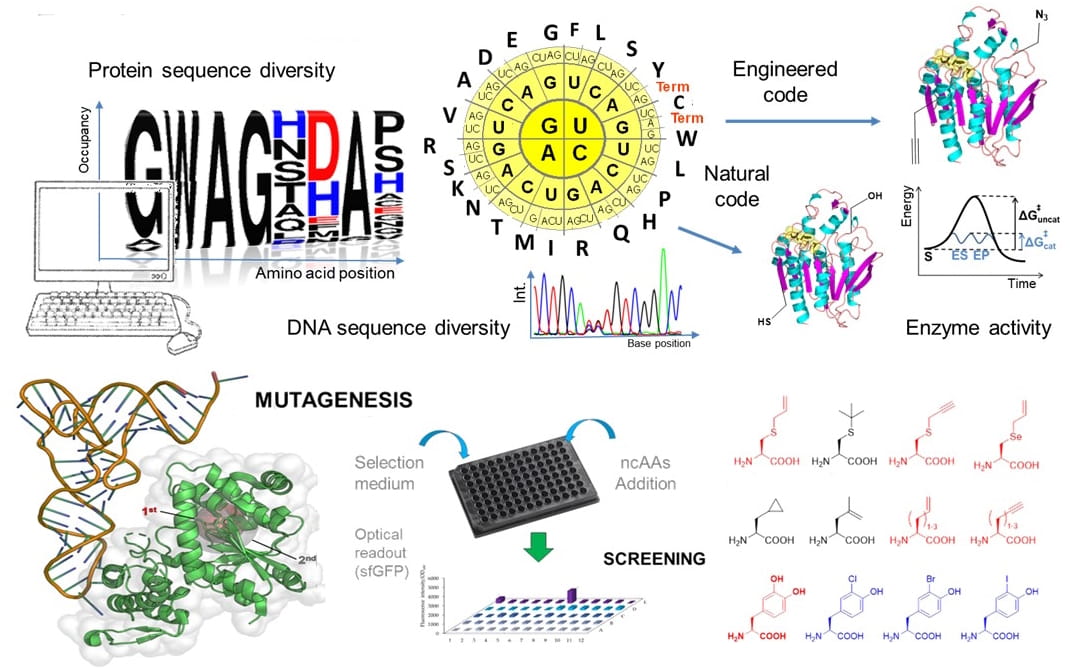
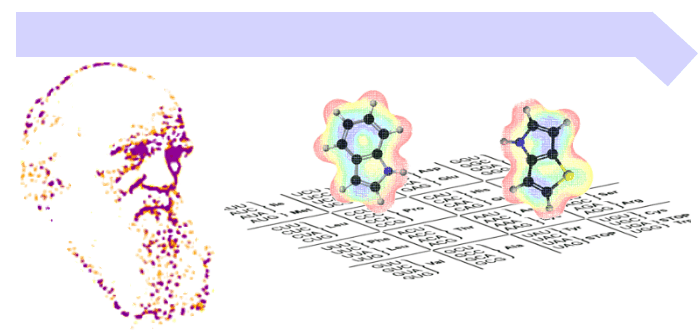
Aminoacyl-tRNA synthetases (aaRSs) play a crucial role in translating the genetic code by linking each amino acid to its matching tRNA. To expand beyond the standard set of 20 amino acids, we modify aaRS specificity, allowing them to “read” new amino acids. By applying directed evolution - often aided by machine learning - we have created several “orthogonal pairs” that boost protein expression. Over the last two decades, we have built a pipeline to design these pairs for diverse applications. We are currently focusing on the directed evolution of dedicated enzymes for small aliphatic and aromatic ncAAs for various ongoing internal and collaborative projects.
Reading for more in-depth coverage of the subject
- Hauf, M., Richter, F., Schneider, T., Faidt, T., Martins, B. M., Baumann, T., Durkin, P., Dobbek, H., Jacobs, K., Moeglich, A. and Budisa, N. (2017) Photoactivatable mussel-based underwater adhesive proteins by an expanded genetic code. ChemBioChem, 18, 1819–1823; doi: 10.1002/cbic.201700327
- Baumann, T.; Hauf, M.; Richter, F.; Albers, S.; Möglich, A.; Ignatova, Z.; Budisa, N. (2019). Computational Aminoacyl-tRNA Synthetase Library Design for Photocaged Tyrosine. Int. J. Mol. Sci. 20 (9), e2343; doi: 10.3390/ijms20092343
Adaptive Laboratory Evolution (ALE) of Escherichia coli

We strive to create synthetic cells using a top-down synthetic biology approach, engineering Escherichia coli to thrive on synthetic amino acids and even plastics as their sole carbon source. In doing so, these biosafe cells will generate protein-based biomaterials vital for tissue engineering, materials science, and environmental remediation. Our ALE experiments focus on developing auxotrophic E. coli strains that can metabolize halogenated amino acids or safely produce high-value synthetic compounds - such as peptide-based drugs and bioadhesive materials - from readily available carbon sources like synthetic plastics.
Reading for more in-depth coverage of the subject
- Treiber-Kleinke, C., Berger, A.A., Adrian, L., Budisa, N., Koksch, B., (2024) Escherichia coli adapts metabolically to 6- and 7-fluoroindole, enabling proteome-wide fluorotryptophan substitution. Front. Synth. Biol. 1:1345634; doi: 10.3389/fsybi.2023.1345634
- Tolle, I., Oehm, S., Hoesl, M.G., Treiber-Kleinke, C., et al. & Rappsilber, J. Ignatova, Z., Gerstein, A.C. Budisa, N. (2023) Evolving a mitigation of the stress response pathway to change the basic chemistry of life. Front. Synth. Biol. 1:1248065; doi: 10.3389/fsybi.2023.1248065